
Our new paper, published in Microbial Genomics, highlights a successful collaboration between microbiologists and HUGS parasitologists. We collected snail faeces across Africa and the UK, identifying a reservoir of antimicrobial resistance (AMR) genes.
We recently proposed a rationale for connecting AMR surveillance with schistosomiasis research. This approach not only optimises sample usage but also reflects the interconnected nature of parasite and AMR transmission, especially in regions with poor water, sanitation and hygiene (WASH) infrastructure.
The HUGS team has routinely monitored freshwater snail populations in Malawi over the last three years to study environmental schistosome transmission. At these sites, human and animal water contact is a common sight, prompting us to question whether these snails might also serve as reservoirs of AMR bacteria. Such a connection could arise from enteric bacteria introduced into the habitat or from environmental AMR propagators lurking in the water.
To investigate this, we initially collected snail faeces from six sites in Malawi. We later expanded the study, incorporating samples collected in the United Kingdom, Zanzibar (in collaboration with the Ministry of Health), and Uganda (in collaboration with the Vector Control Division). The DNA extracted from these samples underwent metagenomic sequencing, with data analysis conducted in Adam Roberts’ group at LSTM.
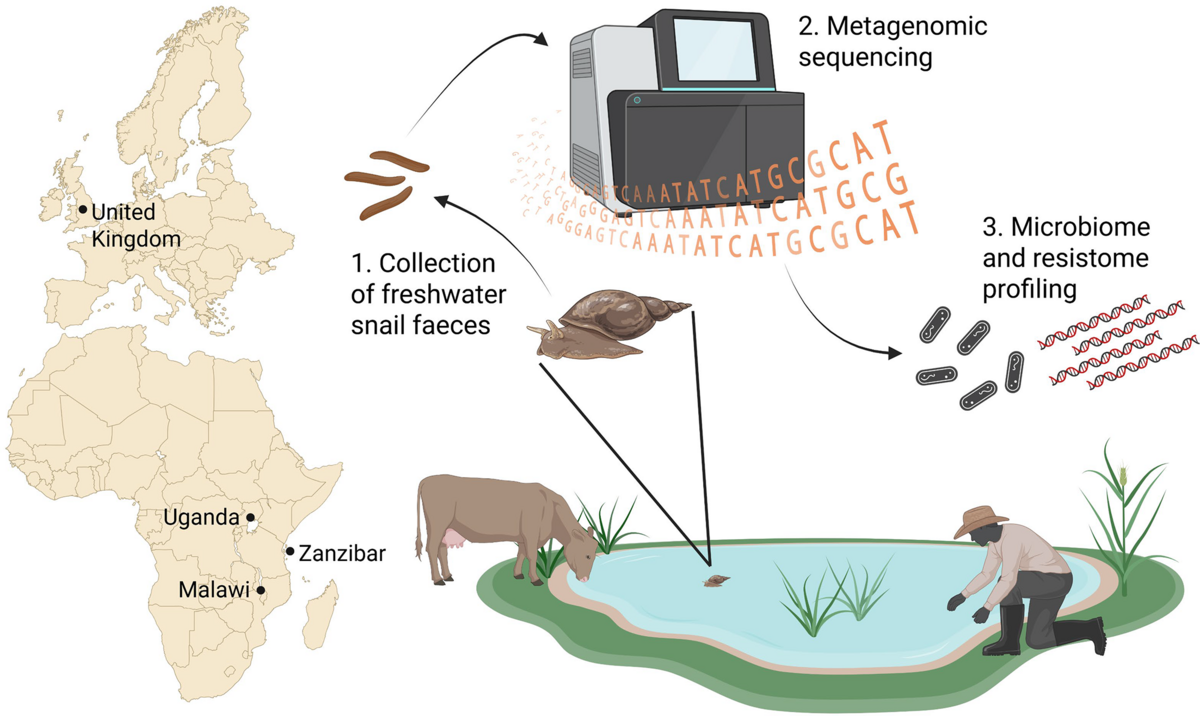
We found that snail faeces are indeed rich in AMR genes and mobile genetic elements associated with both environmental and clinically relevant bacteria. Concerning genes that are predicted to confer resistance to last-resort antibiotics, such as carbapenems and colistin, have been identified. These genes pose a threat of emerging AMR in areas where reliance on last-resort antibiotics may increase due to rising resistance to other available antibiotic treatments.
Our findings reinforce the importance of integrating One Health principles into AMR surveillance and bolster arguments for strengthening WASH infrastructure globally. By linking this work to our human and animal work, we aim to build a compelling case for interconnected pathogen surveillance systems and hope to advocate for streamlined, efficient solutions that tackle these pressing global health challenges.